Overview
A machine learning project focused on mushroom classification using multiple models and comparing their performance. The project includes comprehensive data preprocessing, feature engineering, and model evaluation using cross-validation techniques.
Data Processing Pipeline
Data Preparation
- Loading and splitting semi-colon separated dataset
- Handling 21 different features including:
- Categorical: cap shape, surface, color, gill properties
- Continuous: stem dimensions, cap diameter
- Target variable: mushroom classification
Feature Engineering
-
Categorical Features Processing:
- One-hot encoding for 17 categorical features
- Feature label generation
- Conversion to sparse matrix format
-
Continuous Features Processing:
- Standard scaling for numerical features
- Normalization of stem dimensions and cap diameter
- Feature concatenation for final dataset
Model Implementation
Cross-validation
- Implemented k-fold cross-validation (k=10)
- Consistent evaluation across all models
- Robust performance measurement
Models Evaluated
-
K-Nearest Neighbors (KNN)
- Tested k values from 1 to 10
- Best performance at k=15
- Accuracy: 100%
-
Support Vector Machine (SVM)
- Linear kernel implementation
- C=1 hyperparameter
- Accuracy: 88%
-
Decision Tree
- Default parameters
- Accuracy: 100%
-
Gaussian Naive Bayes
- Probabilistic classifier
- Accuracy: 60%
-
Neural Network
- MLP Classifier
- Alpha=1, max_iterations=1000
- Accuracy: 100%
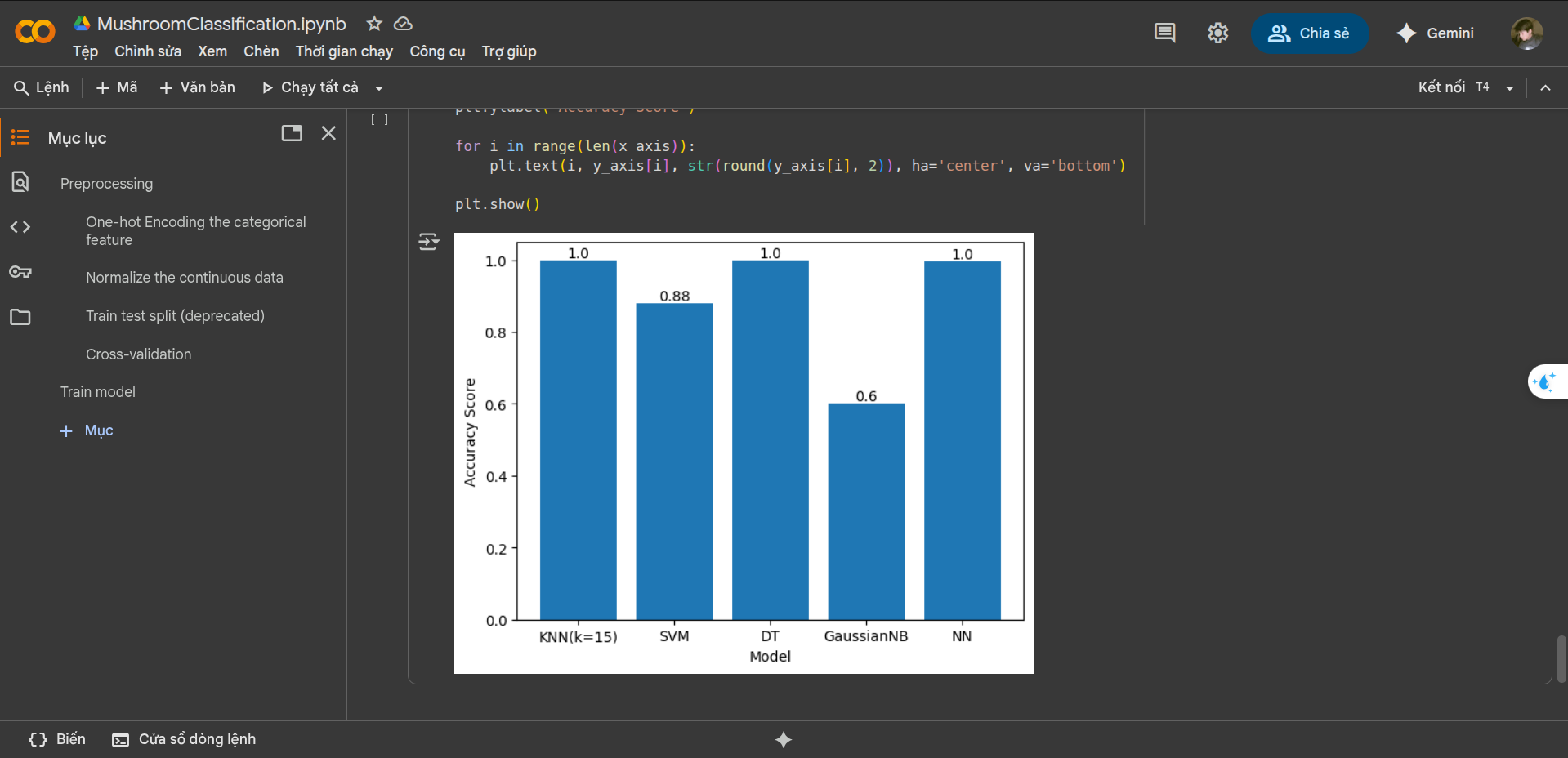
Results Analysis
Performance Comparison
- Top Performing Models:
- KNN (k=15): 100%
- Neural Network: 100%
- Decision Tree: 100%
- Mid-Range Performance:
- SVM: 88%
- Lower Performance:
- Gaussian NB: 60%
Key Findings
- Three models achieved perfect classification
- SVM showed good but not optimal performance
- Gaussian NB struggled with the feature space
Technical Implementation Details
Libraries Used
- Pandas for data manipulation
- Scikit-learn for ML models
- NumPy for numerical operations
- Matplotlib for visualization
Code Structure
- Modular preprocessing pipeline
- Cross-validation implementation
- Model training and evaluation
- Performance visualization
Future Improvements
- Feature importance analysis
- Hyperparameter tuning
- Ensemble methods implementation
- Additional data collection
- Model optimization for speed